STA 225 2.0 Design and Analysis of Experiments
Lecture 7 - 9
Dr Thiyanga S. Talagala
2021-11-16/ 2021-11-19/ 2021-11-26
Completely randomized design
Advantages of CRD
Easy to design
Analysis of data is simple and straight forward
Even if some values are missing the analysis can be done
Disadvantages of CRD
The design requires homogeneous set of experimental units
If the experimental units are not homogeneous, error component will be large and this will make the treatment comparison less efficient.
Randomized Complete Block Design (RCBD)
Suppose we want to compare rr treatments
To run a CRD, we need to find n1+n2+....+nrn1+n2+....+nr homogeneous experimental units to apply treatment.
Often it is difficult to find enough experimental homomogeneous units.
However, it may be possible to find several blocks of experimental units, with enough homogeneous experimental units to apply rr treatments.
RCBD can be used to compare treatment means in such situations even though there are differences between blocks.
Nuisance factor
A factor that has an effect on the response, but we are not interested in that effect.
- Unknown and uncontrollable: Randomization to balance out its effect
Known and uncontrollable but measurable: Analysis of covariance (ANCOVA) (at the time of the analysis)
Nuisance source of variability is Known and controllable: use Blocking to systematically eliminate its effect on the statistical comparison among treatment means.
Example
We want to test the difference between different fertilizers (A, B, C, D, E, F) on strawberry yield. The experimenter has decided to obtain four observations for each fertilizer type.
Example
We want to test the difference between different fertilizers (A, B, C, D, E, F) on strawberry yield. The experimenter has decided to obtain four observations for each fertilizer type. How many experimental units are required to compare the treatments?

Example (cont.)
The experiment is run at 4 different locations having 6 different plots of land each. Hence, a block is given by a location and an experimental unit by a plot of land.
Example (cont.)
randomized complete block design (RCBD)
the experimental units are homogeneous within a block
within each block, the 6 treatments are randomly assigned to 6 experimental units
each block (farms) contains all the treatments (fertilizer)
the design is called complete because we see the complete set of treatments within every block
RCBD
In-class diagram
RCBD
Randomization:
Number the aa treatments 1,2,…,a1,2,…,a.
First form the homogeneous blocks of the experimental units. Then allocate each treatment randomly in each block.
- Number the units in each block as 1,2,...,a1,2,...,a.
- Randomly allocate the aa treatments to aa experimental units in each block
RCBD
Replication
Since each block contains all the treatments, so every treatment will appear in all the blocks. So each treatment can be considered as if replicated the number of times as the number of blocks. Hence, in RCBD, the number of blocks and the number of replications are same.
Your turn
Question: How the 4 diets (A, B, C, and D) affect the coagulation of rabbits?
Treatment: Diet
Factor levels: A, B, C, D
Response: Time in seconds that it takes for a cut to stop bleeding (coagulation rate).
Experimental unit: 16 rabits
Replicates: 4
There are 16 rabbits (same age, weight, height)
Your turn
Which approach do you use to analyze the data? CRD or RCBD
Have a jar with the letters A, B, C, D written on separate slips. Catch a rabbit, pick a slip at random from the bowl and assign the rabbit to the diet letter on the slip. Do not replace the slip. Catch the second rabbit and select another slip from the remaining three slips. Assign that treatment to the second rabbit. Continue until the first four rabbits are assigned one of the four diets. Replace the slips and repeat the procedure until all rabbits are assigned to a diet.
Which approach do you use to analyze the data? CRD or RCBD
source: here
Notations
| Block 1 | Block 2 | ... | Block b |
|---|---|---|---|
| y11y11 | y12y12 | ... | y1by1b |
| y21y21 | y22y22 | ... | y2by2b |
| . | |||
| . | |||
| . | |||
| ya1ya1 | ya2ya2 | ... | yabyab |
Number of treatments: aa
Number of blocks: bb
"There is one observation per treatment in each block, and the order in which the treatments are run within each block is determined randomly. Because the only, randomization of treatment is within the blocks, we often say that the blocks represent a restriction on randomization". (Montgomery, Design and Analysis of Experiments, 2001)
Statistical Model for the RCBD
1. Mean model
Yij=μij+ϵij{i=1,2,...,aj=1,2,...,b
a - number of treatments
b - number of blocks
μij - mean of the ith factor level or treatment and jth block
ϵij - random error (It is assumed ϵij's are independent and N(0,σ2))
Let
μ - overall mean,
τi - effect of the ith treatment,
βj - effect of the jth block,
Then, μij=μ+τi+βj.
Statistical Model for the RCBD
2. Effects model
Yij=μ+τi+βj+ϵij{i=1,2,...,aj=1,2,...,b
a - number of treatments
b - number of blocks
μ - overall mean
τi - effect of the ith treatment
βj - effect of the jth block
ϵij - random error with NID(0,σ2)
Two constraints
Treatment effect and block effects are the deviations from the overall mean. Hence
a∑i=1τi=0 and
b∑j=1βj=0
In-class
In CRD
Yij=μ+τi+ϵij{i=1,2,...,aj=1,2,...,n
τi=μ+μi, i=1,2,...a
∑ai=1μia=μ
This definition implies
a∑i=1τi=0
In-class
Hypothesis - RCBD
We want to test the equality of the treatment means
H0:μ1=μ2=...=μa H1:at least one μi≠μj
The ith treatment mean can be written as
μi=∑bj=1(μ+τi+βj)b=μ+τi. Hence, an equivalent way of writing the above hypothesis
H0:τ1=τ2=...=τa=0 H1:τi≠0 at least one i
Notations
yi. - total of all observations taken under treatment i
Write the mathematical equation
y.j - total of all observations in block j
Write the mathematical equation
y.. - the grand total of all observations
Write the mathematical equation
N=ab be the total number of observations
Notations
ˉyi. - the average of the observations taken under treatment i
ˉy.j - the average of the observations in block j
ˉy.. - the grand average of all observations
ˉyi.=yi.b
ˉy.j=y.ja
ˉy..=y..N
We express the total corrected sum of squares
a∑i=1b∑j=1(yij−ˉy..)2=a∑i=1b∑j=1[(ˉyi.−ˉy..)+(ˉy.j−ˉy..)+(yij−ˉyi.−ˉy.j+ˉy..)]2
Note:
The sum of squares is the sum of the squared values of a variable.
For example the total corrected sum of squares is the sum of the squared values after subtracting (i.e. correcting for) their mean.
Your turn
a∑i=1b∑j=1(yij−ˉy..)2=a∑i=1b∑j=1[(ˉyi.−ˉy..)+(ˉy.j−ˉy..)+(yij−ˉyi.−ˉy.j+ˉy..)]2
Show that the above can be simplified into
a∑i=1b∑j=1(yij−ˉy..)2=ba∑i=1(ˉyi.−ˉy..)2+ab∑j=1(ˉy.j−ˉy..)2+a∑i=1b∑j=1(yij−ˉy.j−ˉyi.+ˉy..)
SST=SSTreatment+SSBlock+SSE
ANOVA Table
| Source of variation | Sum of squares (SS) | DF | Mean Square (MS) | F | p-value | |
|---|---|---|---|---|---|---|
| Treatments | SSTreatments | a−1 | MSTreatments | F0=MSTreatmentsMSE | P(F≥F0) | |
| Blocks | SSBlocks | b−1 | MSBlocks | |||
| Error | SSE | (a−1)(b−1) | MSE | |||
| Total | SST | N−1 |
Example
Section 4.1, Montgomery, D. C. (2017). Design and analysis of experiments. John wiley & sons.
Experiment: Hardness testing experiment
We wish to determine whether 4 different tips produce different (mean) hardness reading on a hardness tester.
A hardness testing machine operates by pressing a tip into a metal test “coupon.”
The hardness of the coupon is measured from the depth of the resulting depression. - Response variable
Four tip types are being tested to see if they produce significantly different readings. - Treatment
Example (cont.)
The coupons might differ slightly in their hardness (for example, if they are taken from ingots produced in different heats). - Block
Within each coupon (block) the order in which the four tips were tested was randomly determined.
References: https://web.ma.utexas.edu/users/mks/384Esp08/rcbdexample.pdf
Example (cont.)
tip <- c(1, 2, 3, 4)coupon1 <- c(9.3, 9.4, 9.2, 9.7)coupon2 <- c(9.4, 9.3, 9.4, 9.6)coupon3 <- c(9.6, 9.8, 9.5, 10.0)coupon4 <- c(10.0, 9.9, 9.7, 10.2)df <- data.frame(tip=tip, coupon1=coupon1, coupon2=coupon2, coupon3=coupon3, coupon4=coupon4)df tip coupon1 coupon2 coupon3 coupon41 1 9.3 9.4 9.6 10.02 2 9.4 9.3 9.8 9.93 3 9.2 9.4 9.5 9.74 4 9.7 9.6 10.0 10.2summary(df[, 2:5]) coupon1 coupon2 coupon3 coupon4 Min. :9.200 Min. :9.300 Min. : 9.500 Min. : 9.70 1st Qu.:9.275 1st Qu.:9.375 1st Qu.: 9.575 1st Qu.: 9.85 Median :9.350 Median :9.400 Median : 9.700 Median : 9.95 Mean :9.400 Mean :9.425 Mean : 9.725 Mean : 9.95 3rd Qu.:9.475 3rd Qu.:9.450 3rd Qu.: 9.850 3rd Qu.:10.05 Max. :9.700 Max. :9.600 Max. :10.000 Max. :10.20library(tidyverse)df.pl <- df %>% pivot_longer(2:5, "Block", "value")df.pl# A tibble: 16 × 3 tip Block value <dbl> <chr> <dbl> 1 1 coupon1 9.3 2 1 coupon2 9.4 3 1 coupon3 9.6 4 1 coupon4 10 5 2 coupon1 9.4 6 2 coupon2 9.3 7 2 coupon3 9.8 8 2 coupon4 9.9 9 3 coupon1 9.210 3 coupon2 9.411 3 coupon3 9.512 3 coupon4 9.713 4 coupon1 9.714 4 coupon2 9.615 4 coupon3 10 16 4 coupon4 10.2library(tidyverse)df.pl$tip <- as.factor(df.pl$tip)df.block <- df.pl %>% group_by(Block) %>% summarize(mean = mean(value))df.block# A tibble: 4 × 2 Block mean <chr> <dbl>1 coupon1 9.4 2 coupon2 9.433 coupon3 9.724 coupon4 9.95df.treatment <- df.pl %>% group_by(tip) %>% summarize(mean = mean(value))df.treatment# A tibble: 4 × 2 tip mean <fct> <dbl>1 1 9.572 2 9.6 3 3 9.454 4 9.88Block
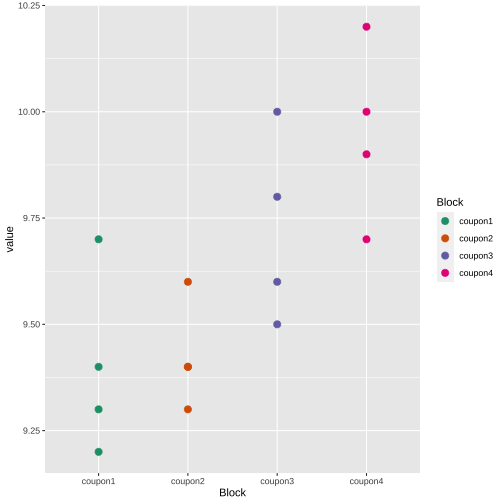
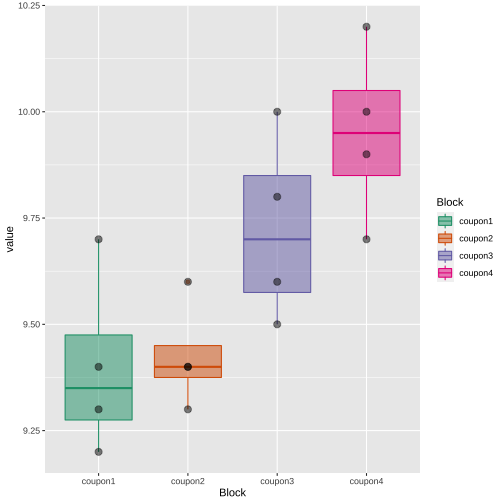
Treatment
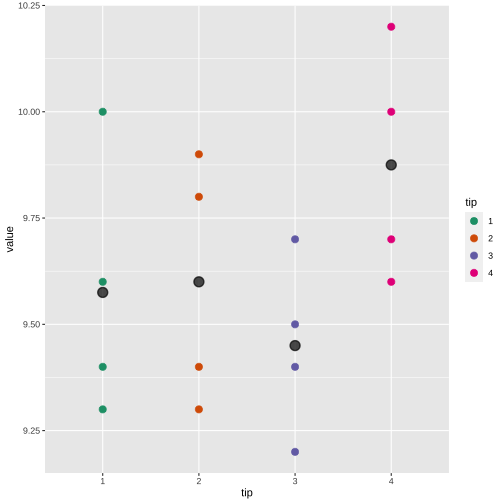
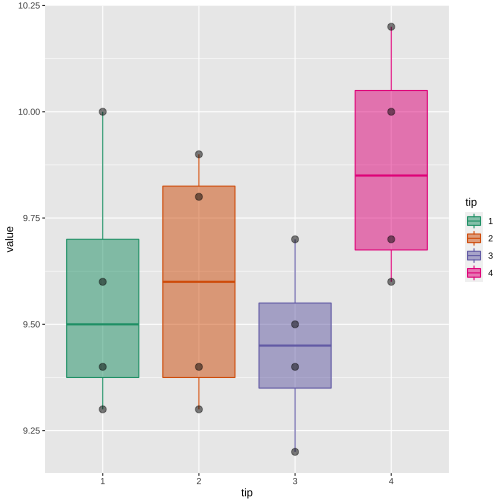
ANOVA
two.way <- aov(value~ tip + Block, data = df.pl)summary(two.way) Df Sum Sq Mean Sq F value Pr(>F) tip 3 0.385 0.12833 14.44 0.000871 ***Block 3 0.825 0.27500 30.94 4.52e-05 ***Residuals 9 0.080 0.00889 ---Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Model Adequacy Checking
Residuals
library(broom)residdf <- augment(two.way)Warning: Tidiers for objects of class aov are not maintained by the broom team,and are only supported through the lm tidier method. Please be cautious ininterpreting and reporting broom output.residdf# A tibble: 16 × 9 value tip Block .fitted .resid .hat .sigma .cooksd .std.resid <dbl> <fct> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> 1 9.3 1 coupon1 9.35 -0.0500 0.438 0.0972 5.56e- 2 -0.707 2 9.4 1 coupon2 9.38 0.0250 0.438 0.0993 1.39e- 2 0.354 3 9.6 1 coupon3 9.67 -0.0750 0.438 0.0935 1.25e- 1 -1.06 4 10 1 coupon4 9.9 0.100 0.438 0.0882 2.22e- 1 1.41 5 9.4 2 coupon1 9.38 0.0250 0.437 0.0993 1.39e- 2 0.354 6 9.3 2 coupon2 9.4 -0.100 0.438 0.0882 2.22e- 1 -1.41 7 9.8 2 coupon3 9.7 0.100 0.437 0.0882 2.22e- 1 1.41 8 9.9 2 coupon4 9.93 -0.0250 0.437 0.0993 1.39e- 2 -0.354 9 9.2 3 coupon1 9.22 -0.0250 0.438 0.0993 1.39e- 2 -0.35410 9.4 3 coupon2 9.25 0.150 0.438 0.0707 5.00e- 1 2.12 11 9.5 3 coupon3 9.55 -0.0500 0.438 0.0972 5.56e- 2 -0.70712 9.7 3 coupon4 9.78 -0.0750 0.438 0.0935 1.25e- 1 -1.06 13 9.7 4 coupon1 9.65 0.0500 0.438 0.0972 5.56e- 2 0.70714 9.6 4 coupon2 9.68 -0.0750 0.438 0.0935 1.25e- 1 -1.06 15 10 4 coupon3 9.98 0.0250 0.437 0.0993 1.39e- 2 0.35416 10.2 4 coupon4 10.2 0 0.438 0.100 2.81e-30 0The normality assumption
ggplot(residdf, aes(x=.resid))+ geom_histogram(colour="white")+ggtitle("Distribution of Residuals")`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
ggplot(residdf, aes(sample=.resid))+ stat_qq() + stat_qq_line()+labs(x="Theoretical Quantiles", y="Sample Quantiles")
The normality assumption (cont.)
shapiro.test(residdf$.resid) Shapiro-Wilk normality testdata: residdf$.residW = 0.93957, p-value = 0.3438Plot of the residuals vs type
ggplot(data=residdf, aes(x=tip, y=.resid)) + geom_point()
Plot of the residuals vs block
ggplot(data=residdf, aes(x=Block, y=.resid)) + geom_point()
Plot of residuals versus fitted values
To check the assumption of constant variance
Residuals should be structureless. Residuals should not contain any obvious patterns
ggplot(data=residdf, aes(x=.fitted, y=.resid)) + geom_point()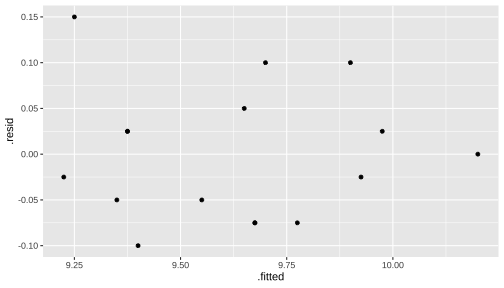
ANOVA - Computing formulas
SST=a∑i=1b∑j=1y2ij−y2..N SSTreatments=1ba∑i=1y2i.−y2..N
SSBlocks=1ab∑i=j=1y2.j−y2..N
SSE=SST−SSTreatments−SSBlocks
Expected value of mean squares, if treatments and blocks are fixed
E(MSTreatments)=σ2+b∑ai=1τ2ia−1
E(MSBlocks)=σ2+a∑bj=1β2jb−1
E(MSE)=σ2
Test the equality of treatment means
F0=MSTreatmentsMSE which is distributed as Fa−1,(a−1)(b−1)
Comparing block treatment means
F0=MSBlocksMSE which is distributed as Fb−1,(a−1)(b−1)
RCBD - Multiple Comparisons
The rejection of the null hypothesis indicates a significant difference in treatment means.
Any of the multiple comparison methods can be used for detecting which means are significantly different. In this course we use, Tukey's method can be used.
In general we are not interested to compare block means.
TukeyHSD(two.way, "tip") Tukey multiple comparisons of means 95% family-wise confidence levelFit: aov(formula = value ~ tip + Block, data = df.pl)$tip diff lwr upr p adj2-1 0.025 -0.18311992 0.23311992 0.98090053-1 -0.125 -0.33311992 0.08311992 0.30275634-1 0.300 0.09188008 0.50811992 0.00665833-2 -0.150 -0.35811992 0.05811992 0.18159074-2 0.275 0.06688008 0.48311992 0.01132844-3 0.425 0.21688008 0.63311992 0.0006061plot(TukeyHSD(two.way, "tip"))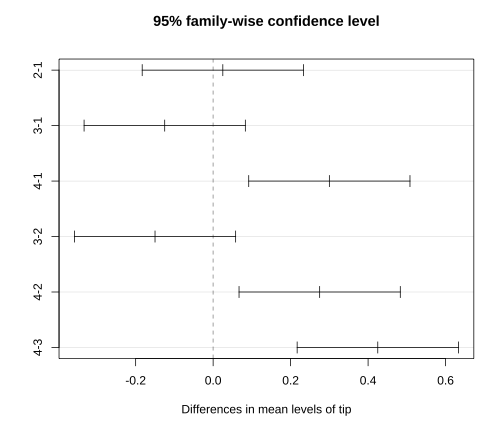
Randomized Incomplete Block Designs
Every treatment is not present in every block
Balanced Incomplete Block Design (BIBD)
All pairs of treatments occur together within a block an equal number of times
Acknowledgement
Some of the slide content is based on
Montgomery, D. C. (2017). Design and analysis of experiments. John wiley & sons.